These Creatures Are Helping Us Understand Our Genome
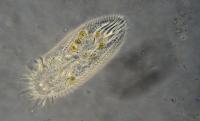
An Oxytricha cell swimming in pond water. Image courtesy of Graham Matthews.
A new study of a single-celled eukaryote with 16,000 tiny chromosomes may shed light on a recently discovered feature of the human genome.
Methyladenine, or 6mA—a modification of DNA common in Oxytricha trifallax—has only recently been found in multicellular organisms, with some studies suggesting a role in human disease and development.
Finding the enzymes that lay down the methyl marks will be critical to understanding what 6mA is doing in Oxytricha and other organisms, but the enzymes have been difficult to identify.
The new research—to be published in the June 13 issue of Cell—reveals how 6mA marks are made to the Oxytricha genome and suggests why the enzymes have been hard to find.
Not Your Ordinary Model Organism

Based on appearances, Oxytricha trifallax doesn’t look like an ordinary model organism for biomedical research.
With a fringe of exterior cilia, Oxytricha swims in ponds and collects algae to eat. It does not cause any human diseases.
And then there’s its genome, for which the words “shattered,” “scrambled,” flipped,” and “encrypted” are routine descriptors. At one stage of its life, an Oxytricha cell breaks its genome into a quarter million pieces, swaps information with another cell, tosses out what it doesn’t need, and then stitches everything together into a new working genome.
But the Oxytricha genome has a more conventional side as well. Yes, it has more than 16,000 chromosomes so small they are called nanochromosomes—most just big enough to encode one gene—but each nanochromosome has almost all the same features as humans, mice, and other animals studied by medical researchers.
“Except for the lack of centromeres, they have everything we have—including telomeres, histones, and base modifications—so they provide elegant models for the study of chromatin biology and how DNA is packaged,” says the study’s senior author Laura Landweber, PhD, professor of biochemistry & molecular biophysics and of biological sciences at Columbia University, who has been studying Oxytricha for two decades and previously uncovered Oxytricha’s 16,000 chromosomes.
The Mystery of 6mA
Until recently, DNA N6 adenine methylation (6mA for short) had only been seen in bacteria and single-celled eukaryotes.
“Even though 6mA was discovered in Oxytricha in the '70s, it wasn’t studied much,” says Leslie Beh, PhD, who’s been investigating 6mA in the Landweber lab at Columbia and with Tom Muir, PhD, at Princeton University.
“But in the last four years, there has been a renaissance in 6mA studies as new techniques are showing that it may also be found in Drosophila, C. elegans, mice, and perhaps in humans. There have been some papers that link 6mA to cancer and brain function, so there’s a lot of interest now in what these marks are doing in multicellular organisms.”
To understand what the 6mA marks are doing in Oxytricha, Beh and Landweber first had to identify the enzyme that lays down the marks.
Finding 6mA enzymes has proved to be tricky in other organisms—hampering research into the function of these marks—and Beh’s finding suggests why.
6mA Marks Made By a Complex
Oxytricha uses not one protein, he found, but a complex of four proteins to mark DNA, including an enzyme that originally existed to mark RNA. “The twist is that Oxytricha has recruited an enzyme that acts on RNA and turned it into one that acts on DNA, but it did not follow the simple evolutionary step of turning the RNA binding domain into a DNA binding domain,” Beh says. “Instead it has recruited two partner proteins that have the needed DNA binding characteristics.”
Landweber suspects that other organisms may have done the same, and the finding may help researchers find other, still hidden 6mA enzymes. “Phylogenetic analysis showed that relatives of this enzyme complex are only found in single-celled eukaryotes, but the principle is probably going to extend elsewhere,” she says.
6mA Disfavors Nucleosomes

In Oxytricha, 6mA modified bases (yellow) always occur between nucleosomes, where DNA wraps around histones (turquoise), and may be pushing the nucleosomes away. Magenta on the chromosome ends represents telomeres. Illustration by Myles Marshall.
Beh’s experiments with synthetic Oxytricha chromosomes suggest 6mA is contributing to chromatin organization by controlling where nucleosomes are located. In Oxytricha, 6mAs are always located between nucleosomes, and Beh’s in vitro experiments—which placed 6mA in alternative locations on the chromosome—suggest 6mA may be pushing nucleosomes away from the marks.
This means 6mA could potentially be connected to human diseases, since the alteration of 6mA patterns may lead to the disruption of chromatin organization in essential genes.
6mA Loss Disrupts Oxytricha Genome
By knocking out the catalytic subunit of the complex’s genes, Beh and Landweber also created Oxytricha cells incapable of making 6mA modifications to DNA.
The changes don’t seem to have a big impact on the cell until it tries to exchange DNA with another Oxytricha cell. The exchange happens, but the cell can’t complete the process and produce a working genome.
“The cells die, and we need to figure out why,” Landweber says. “We need to find which step in development is dependent on 6mA methylation. It could be something simple; maybe a certain gene needs this base modification. Or it could be that 6mA is critical to the whole genome remodeling cascade that takes place in these cells.”
And understanding the genome remodeling Oxytricha does could also eventually provide insights into cancer.
“The cells go through genome rearrangements similar to cancer cells, except in reverse,” Landweber says. “In cancer, the genome often gets messed up; in Oxytricha, it starts out messed up and then it’s put back together. There are a lot of shared mechanisms, even though they produce different products in the end.”
References
The study, titled “Identification of a DNA N6-adenine methyltransferase complex and its impact on chromatin organization," first appeared online May 16 in Cell.
Other authors: Galia T. Debelouchina (Princeton University), Derek M. Clay (Columbia University Irving Medical Center), Robert E. Thompson (Princeton), Kelsi A. Lindblad (CUIMC), Elizabeth R. Hutton (Princeton), John R. Bracht (American University, Washington, DC), Robert P. Sebra (Icahn School of Medicine, New York), and Tom W. Muir (Princeton).
This work was funded by NIH grants R01GM59708, R35GM122555, R01GM109459, and R01GM107047.
The authors declare no conflicts of interest.